Multi-omics studies of spider silk (Aachen, King’s College Presentation Part 2)
[Bold text contains links]
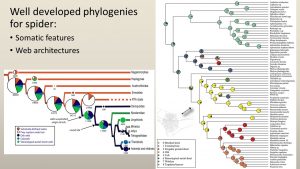
There are now well-developed phylogenies tracking the evolution of spider webs using genomic and phylogenomic information (such as those below). This is great, as we now can easily discern trends, such as the repeated loss and gain of the orb web, that were not able to be gleaned from any web structure analyses.
When it comes to silk types, and their properties, and features such as nano-surfaces and adhesiveness, virtually nothing is known across spider phylogenies. We thus do not know how and why certain nano-scaled features appeared and their evolutionary significance.
There have been a few recent silk genome and transcriptome studies and these are showing some really interesting results regarding the protein chemistry of the spider silks suite. For instance a paper authored by Babb et al in Nature Genetics revealed that the golden orb weaver Trihinephila clavipes produces a much more complex array of spidroins across all of its silk glands, although certain glans do specialize. For example, there may be 8 or more MaSp proteins and these are expressed in variable amounts across all of the silk glands and even venom glands.
This is important information, but it does complicate the story regarding how silk properties have evolved. What may be more important nonetheless is the amino acid composition of the silk and not necessarily the proteins. This would mean the presence of some key amino acids is all it may take to induce extensibility or some other property into synthesized silks. I did an analysis back in 2012 with collaborators from Tunghai University, Taiwan, and the University of Akron, USA, that showed across 10 species from 5 genera proline seemed to be a key amino acid, positively correlating with silk extensibility while negatively correlating with modulus and strength in ground state induced silks.
Later, a student of mine, Hamish Craig, tested this again in a paper published in 2020 in Journal of the Royal Society Interface, this time incorporating 85 species from 44 genera and found similar correlations between amino acids like proline and extensibility. The amino acid glycine was influential -having a positive influence on modulus and negative influence on extensibility. Interestingly, Serine proved to be as or more influential than either of these other amino acids despite having no specific characteristic to indicate that it should. We speculated that the influence of serine is a product of it acting as a switch within the C- and N-termini of the proteins to switch on folding, this being highly influential over the protein’s secondary properties.
An even bigger dataset has since been compiled and examined, this time of 1098 species of spider. This database contains extremely fine details of the genetic expression of a range of spider silks. The database was primarily developed by colleagues at RIKEN (and see here) and Keio University in Japan. Hamish and myself collaborated on collecting within Australia and were co-authors of the Science Advances paper).

It is now possible to scan for the effects of specific protein expression on silk properties and protein secondary and other structures. We think this paper will open up some immense possibilities, like enable the targeting of specific genes for building silks with specific properties via recombinants and other genetic engineering technologies.

